Machine learning techniques for image processing

Problems
High-throughput microscopy techniques, such as light-sheet imaging, generate huge amounts of data, often in the terabyte (TB) range. The challenge lies in the ability to manage these images and extract relevant information from the raw data, through the so-called metadata. This challenge is even more important in the imaging of complex tissue structures such as the brain which offer a lot of information from a morphological, chemical or functional point of view.
Technology
Algorithms related to Machine Learning and more specifically Deep Learning are commonly used. Much attention is also given to speeding up the calculation processes for metadata extraction. In many cases, very large samples are analyzed with a large amount of data, the analysis of which can extend over many hours of calculation. It is therefore essential to work on both hardware and software to speed up the execution times of the computation.
At the level of big data management, we developed ZetaStitcher, a tool for the fast alignment of multiple adjacent tiles, and efficient access to arbitrary portions of the complete volume.
Outcomes and impact
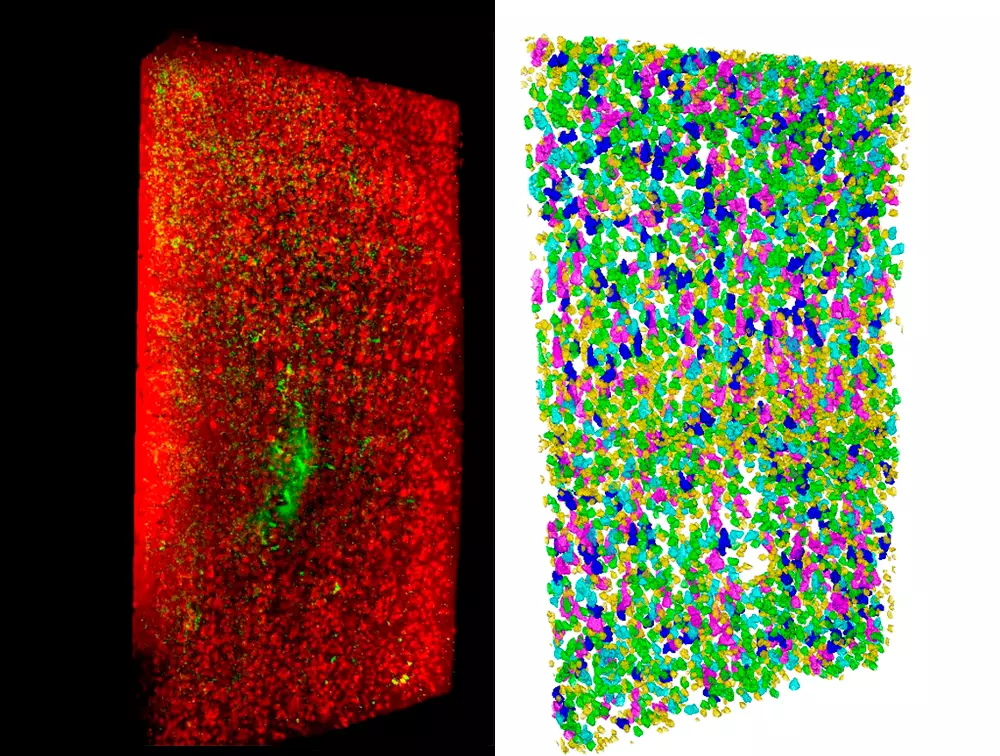
Concerning image analysis, we are exploiting different machine learning strategies to quantify the position, shape and number of fluorescently labelled cells. Semantic deconvolution is used to recover image quality, allowing easier cell detection with standard methods. Deep convolutional neural networks are used to precisely segment neurons, allowing further classification based on cell shape.
One of the main results obtained with these techniques may concern quantitative information about tissues of a cytoarchitectural type, for example:
- the number, spatial position and type of cells
- the orientation of nerve fibres in neural tissue
- physiological data from wearable sensors
More details
Software tools for advanced image processing
High-throughput microscopy techniques, such as light-sheet imaging, generate huge amounts of data, often in the TeraBytes range. The challenge then becomes to manage these images and extract semantically relevant information from the raw data (a large matrix of grayscale values).
At the level of big data management, we developed ZetaStitcher (https://github.com/lens-biophotonics/ZetaStitcher), a tool for fast alignment of multiple adjacent tiles, and for efficient access to arbitrary portions of the complete volume. To orchestrate the operation of our laboratory equipment, we are developing data acquisition and control software in the form of hardware libraries and graphical user interfaces that are able to sustain a high data rate. Further, we are exploring different compression strategies to find the best compromise between data size, image quality preservation, and I/O speed. The tools that we develop are available from our GitHub page (https://github.com/lens-biophotonics).
Concerning image analysis, we are exploiting different machine learning strategies to quantify the position, shape and number of fluorescently labeled cells. Semantic deconvolution is used to recover image quality, allowing easier cell detection with standard methods. Deep convolutional neural networks are used to precisely segment neurons, allowing further classification based on cell shape. All methods are carefully designed to sustain the high data flux of our applications and to require a limited amount of manually annotated ground truth.
Highlights